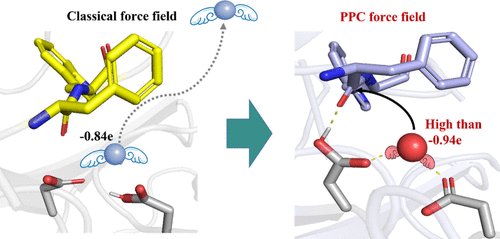
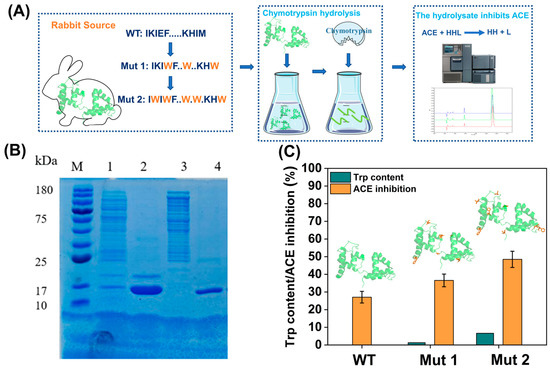
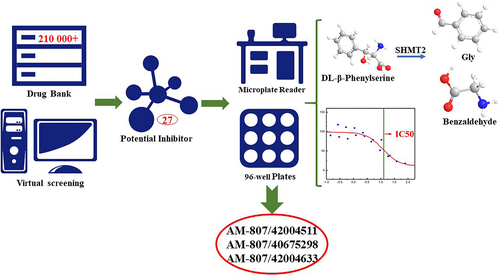
Yalong Cong, Yinghui Feng, Hui Ni, Fengdong Zhi, Yulu Miao, Bohuan Fang, Lujia Zhang,* and John Z.H. Zhang*
COVID-19 has emerged as the most serious international pandemic in early 2020, and the lack of comprehensive knowledge in the recognition and transmission mechanisms of this virus hinders the development of suitable therapeutic strategies. The specific recognition during the binding of the spike glycoprotein (S protein) of coronavirus to the angiotensin-converting enzyme 2 (ACE2) in the host cell is widely considered the first step of infection. However, detailed insights on the underlying mechanism of dynamic recognition and binding of these two proteins remain unknown. In this work, molecular dynamics simulation and binding free energy calculation were carried out to systematically compare and analyze the receptor-binding domain (RBD) of six coronavirus S proteins. We found that affinity and stability of the RBD from SARS-CoV-2 under the binding state with ACE2 are stronger than those of other coronaviruses. The solvent-accessible surface area (SASA) and binding free energy of different RBD subunits indicate an “anchor-locker” recognition mechanism involved in the binding of the S protein to ACE2.Loop2 (Y473–F490) acts as an anchor for ACE2 recognition, and Loop3 (G496–V503) locks ACE2 at the other non-anchoring end. Then, the charged or long-chain residues in the β-sheet1 (N450–F456) region reinforce this binding. The proposed binding mechanism was supported by umbrella sampling simulation of the dissociation process. The current computational study provides important theoretical insights for the development of new vaccines against SARS-CoV-2.